Home
/
Technology
DNBSEQ™ Technology
Discover the Power of DNBSEQ™ Technology
A Revolution in DNA Sequencing
At MGI, we are pioneering a new era of genomic exploration through our innovative DNBSEQ™ technology. By merging cutting-edge engineering with a deep understanding of biology, we empower researchers, clinicians, and industry leaders worldwide to unlock genomic insights with unprecedented accuracy, efficiency, and value.
Reduced Index Hopping & Bias
Circularized templates and RCR minimize error accumulation, improving variant detection accuracy.
Optimized Throughput & Efficiency
Patterned Arrays and precise DNB loading increase signal density, enabling scalable, cost-effective sequencing.
Enhanced Base Calling Precision
Sub-pixel Registration and GPU-accelerated processing deliver faster, more reliable base calls.
High-Fidelity Sequencing
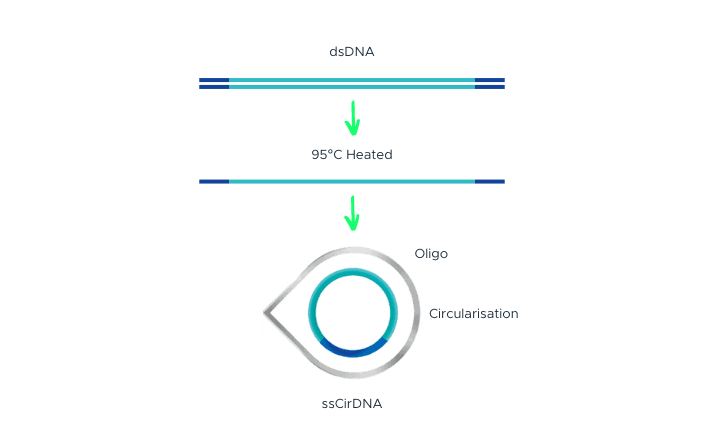
MGI’s DNBSEQ™ technology uses single-strand DNA circularization and rolling circle replication (RCR) to generate DNA Nanoballs (DNBs) directly from the original template.
This approach, combined with Patterned Arrays, cPAS chemistry, and advanced image analysis, reduces index hopping and biases, ensuring more accurate, high-quality sequencing data for both whole-genome and targeted applications.